Creating meshless domains¶
In this tutorial we will see how to:
[1]:
# import necessary libraries
import matplotlib.pyplot as plt
import torch
Some general geometries in 1D, 2D and 3D are already implemented.
[2]:
# import classical domains
from scimba_torch.domain.meshless_domain import (
Cube3D,
Cylinder3D,
Disk2D,
Disk3D,
Polygon2D,
Segment1D,
Square2D,
Torus3D,
TorusFrom2DVolume,
)
Creating a simple Domain¶
To create a simple domain, simply instantiate one of the meshless_domain class with the appropriate parameters.
is_main_domain is used to specify if the domain can have subdomains/holes.[3]:
square = Square2D(
bounds=[(-1, 1), (0, 1)],
is_main_domain=True
)
# to reset the square domain
def simple_square():
return Square2D(
bounds=[(-1, 1), (0, 1)],
is_main_domain=True
)
To visualise a domain, we are going to sample points on it
[4]:
# import necessary libraries
from scimba_torch.integration.monte_carlo import DomainSampler
# a utility function to plot a 2D domain
def plot_2d(domain, n=10_000, ax=None):
"""Plot a 2D domain using a Domain sampler."""
# create the sampler
sampler = DomainSampler(domain)
# sample points
res = sampler.sample(n)
pts = res.x # coordinates of the sampled points
labels = res.labels # labels of the sampled points
# convert to numpy for plotting
pts = pts.detach().cpu().numpy()
labels = labels.detach().cpu().numpy()
# generate the ax if not provided
if ax is None:
fig = plt.figure(figsize=(4, 4))
ax = fig.add_subplot(111, xlabel='x', ylabel='y', aspect='equal')
else:
fig = None
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_aspect('equal')
# plot the points
ax.scatter(pts[:, 0], pts[:, 1], s=1, c=labels, alpha=0.5)
ax.grid(True)
if fig is not None:
fig.tight_layout()
plt.show()
square = simple_square()
plot_2d(square)
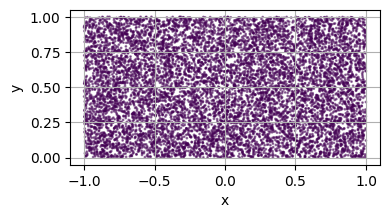
Apply a Mapping to a Domain¶
For more informations about mappings, see Complement on the mapping.
[5]:
from scimba_torch.utils.mapping import Mapping
# define a mapping function
def map(x):
"""A simple mapping function."""
return torch.stack([x[:, 0], x[:, 1]*(torch.abs(x[:, 0]) + 1e-2)], dim=1)
# define the mapping
mapping = Mapping(
from_dim=2,
to_dim=2,
map=map,
)
# set the mapping for the square domain
square = simple_square()
square.set_mapping(
map=mapping,
bounds_postmap=[(-1, 1), (0, 1)],
)
plot_2d(square)
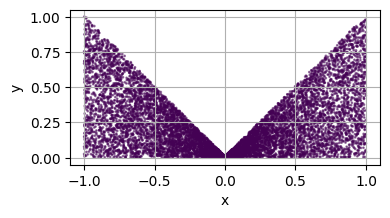
Adding Holes to a Domain¶
is_main_domain set to false.For example, let’s consider a disk hole inside the domain
[6]:
square = simple_square()
# create some holes
hole1 = Disk2D(center=(.8, 0.5), radius=0.1, is_main_domain=False)
hole2 = Disk2D(center=(-.8, 0.5), radius=0.1, is_main_domain=False)
# add holes to the square domain
square.add_hole(hole1)
square.add_hole(hole2)
plot_2d(square)
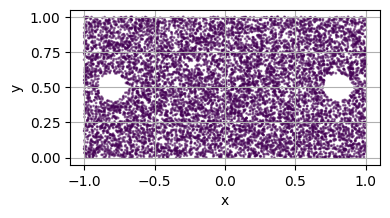
[7]:
# helper for resetting the domains
def simple_disks():
disk_L = Disk2D(center=(-.8, 0.5), radius=0.1, is_main_domain=False)
disk_R = Disk2D(center=(.8, 0.5), radius=0.1, is_main_domain=False)
return disk_L, disk_R
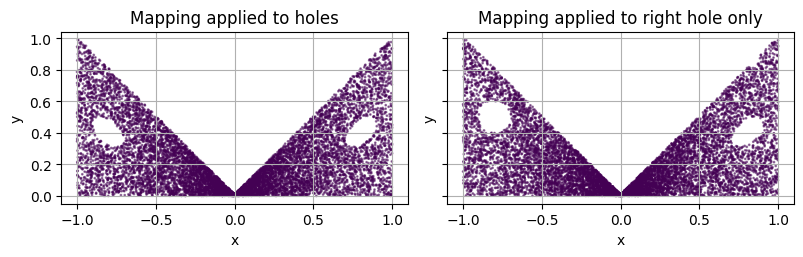
Combining Holes and Mapping¶
We can also combine holes and mappings.
adding a hole (if the mapping is already set), with the flag
copy_mappingset to Falsesetting the mapping for all already added holes, with the flag
to_holesset to False
Yet, we recommend to proceed in the following order:
Create the main domain
Set the mapping if any
Add holes to the domain, with
copy_mappingset to False if we do not want to apply the mapping on the hole
[8]:
fig, axs = plt.subplots(1, 3, figsize=(12, 4), sharex=True, sharey=True)
# by default, the mapping is applied to all the holes
square = simple_square()
disk_L, disk_R = simple_disks()
square.set_mapping(map=mapping, bounds_postmap=[(-1, 1), (0, 1)])
square.add_hole(disk_L)
square.add_hole(disk_R)
# plot
plot_2d(square, ax=axs[0])
axs[0].set_title('Mapping applied to holes')
# we can specify not to apply the mapping to a specific hole when adding it
square = simple_square()
disk_L, disk_R = simple_disks()
square.set_mapping(map=mapping, bounds_postmap=[(-1, 1), (0, 1)])
square.add_hole(disk_L, copy_mapping=False)
square.add_hole(disk_R, copy_mapping=True)
# plot
plot_2d(square, ax=axs[1])
axs[1].set_title('Mapping applied to right hole only')
# # Alternatively, we can specify not to apply the mapping to already existing holes,
# # but we will prefer to set the mapping first and then add the holes
# square = simple_square()
# disk_L, disk_R = simple_disks()
# square.add_hole(disk_L)
# square.set_mapping(map=mapping, bounds_postmap=[(-1, 1), (0, 1)], to_holes=False)
# square.add_hole(disk_R)
# plot2D(square, ax=axs[2])
# axs[2].set_title('Mapping applied to right hole only')
axs[2].remove()
fig.tight_layout()
plt.show()
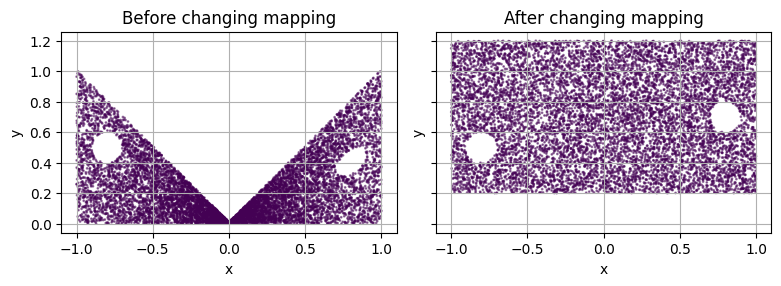
When changing the mapping of the domain (i.e. the domain was already mapped) with the flag to_holes set to False, the mapping will still be applied to previously mapped holes.
[9]:
fig, axs = plt.subplots(1, 2, figsize=(8, 4), sharex=True, sharey=True)
square = simple_square()
disk_L, disk_R = simple_disks()
# define a new mapping
mapping2 = Mapping.translate(torch.tensor([0, 0.2]))
# set the mapping for the square domain
square.set_mapping(map=mapping, bounds_postmap=[(-1, 1), (0, 1)])
# add the holes
square.add_hole(
disk_L, copy_mapping=False
) # left disk hole will NOT have the mapping applied
square.add_hole(disk_R) # right disk hole will have the mapping applied
# plot
plot_2d(square, ax=axs[0])
axs[0].set_title("Before changing mapping")
# apply the new mapping to the square domain
square.set_mapping(map=mapping2, bounds_postmap=[(-1, 1), (0.2, 1.2)], to_holes=False)
# plot
plot_2d(square, ax=axs[1])
axs[1].set_title("After changing mapping")
fig.tight_layout()
plt.show()
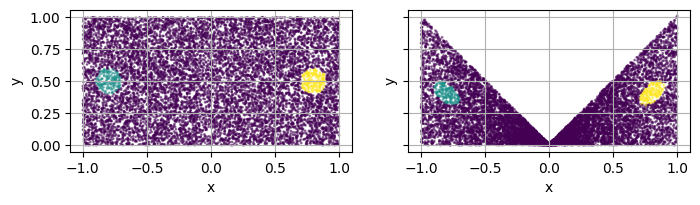
Adding subdomains¶
[10]:
fig, axs = plt.subplots(1, 2, figsize=(8, 4), sharex=True, sharey=True)
square = simple_square()
disk_L, disk_R = simple_disks()
square.add_subdomain(disk_L)
square.add_subdomain(disk_R)
plot_2d(square, ax=axs[0])
square.set_mapping(map=mapping, bounds_postmap=[(-1, 1), (0, 1)])
plot_2d(square, ax=axs[1])
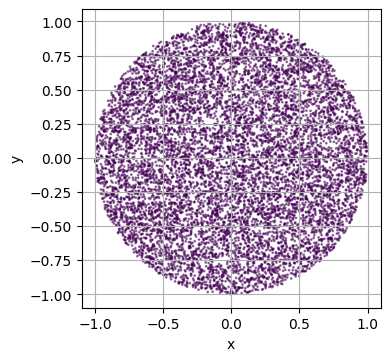
Defining your own Domains with a custom SDF¶
A Signed distance function is simply a Callable with a treshold
[11]:
# import necessary libraries
from scimba_torch.domain.meshless_domain.base import VolumetricDomain
from scimba_torch.domain.sdf import SignedDistance
class CustomSDF(SignedDistance):
"""A custom SDF class that defines the signed distance function for unit 2D disk."""
def __init__(self):
super().__init__(
dim=2,
threshold=0.0,
)
def __call__(self, x: torch.Tensor) -> torch.Tensor:
"""Compute the signed distance function for the unit 2D disk."""
## WARNING, the sdf must return a tensor of shape (N, 1)
x, y = x[:, 0], x[:, 1]
return (torch.sqrt(x**2 + y**2) - 1.0)[:, None] # shape (N, 1)
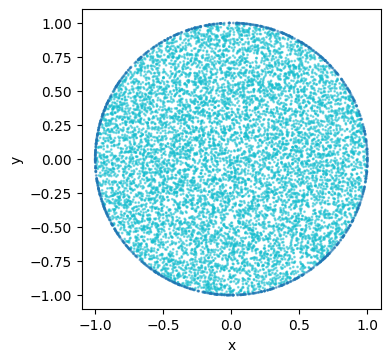
custom_domain = VolumetricDomain(
domain_type="CustomUnitDisk",
dim=2,
sdf=CustomSDF(),
bounds=[(-1, 1), (-1, 1)],
is_main_domain=True
)
plot_2d(custom_domain)
⚠⚠⚠⚠⚠⚠⚠⚠⚠
⚠WARNING⚠: The domain does not have boundary conditions
⚠⚠⚠⚠⚠⚠⚠⚠⚠
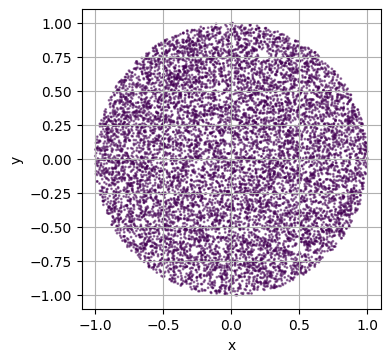
Boundary Domains¶
#TODO : implement default sampling with sdf when no bc domains are given.
As you can see in the last example, our custom domain does not have any boundary domains…
In Scimba boundary domain (more generally called SurfacicDomains), are a “proper” domain (i.e. a VolumetricDomain) of dimension \(d\), mapped to dimension \(d+1\)
To declare such a domain, we hence need:
A
VolumetricDomain(calledparametric_domain)A
Mapping(calledsurface) that embeds ourparametric_domaininto higher dimension.
Let’s define the boundary of our unit disk.
[12]:
# import necessary libraries
from scimba_torch.domain.meshless_domain.base import SurfacicDomain
bc = SurfacicDomain(
domain_type="CustomUnitCircle",
parametric_domain=Segment1D((0, 2 * torch.pi)),
surface=Mapping(
from_dim=1,
to_dim=2,
map=lambda x: torch.stack([torch.cos(x[..., 0]), torch.sin(x[..., 0])], dim=-1),
),
)
# we then add the boundary domain to our custom domain
custom_domain.add_bc_domain(bc)
# we don't have a warning anymore :)
plot_2d(custom_domain)
Let’s create a new plotting utility function to see what’s happening.
[13]:
# a utility function to plot a 2D domain and boundary points
def plot_2d_with_bc(domain, n=10_000, n_bc=1_000, ax=None):
"""Plot a 2D domain using a Domain sampler."""
cmap = plt.get_cmap("tab10_r")
# create the sampler
sampler = DomainSampler(domain)
# sample points
res = sampler.sample(n)
pts = res.x # coordinates of the sampled points
labels = res.labels # labels of the sampled points
res_bc, _ = sampler.bc_sample(n_bc)
pts_bc = res_bc.x # coordinates of the sampled boundary points
labels_bc = res_bc.labels # labels of the sampled boundary points
# convert to numpy for plotting
pts = pts.detach().cpu().numpy()
labels = labels.detach().cpu().numpy()
pts_bc = pts_bc.detach().cpu().numpy()
labels_bc = labels_bc.detach().cpu().numpy()
# generate the ax if not provided
if ax is None:
fig = plt.figure(figsize=(4, 4))
ax = fig.add_subplot(111, xlabel="x", ylabel="y", aspect="equal")
else:
fig = None
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_aspect("equal")
# offset the labels for boundary points and find max label for color mapping
labels_bc += labels.max() + 1
max_label = max(labels.max(), labels_bc.max())
# plot the points
ax.scatter(
pts[:, 0],
pts[:, 1],
s=1,
c=labels,
alpha=0.5,
cmap=cmap,
vmin=0,
vmax=max_label,
)
# plot the boundary points
ax.scatter(
pts_bc[:, 0],
pts_bc[:, 1],
s=1,
c=labels_bc,
cmap=cmap,
vmin=0,
vmax=max_label,
alpha=0.5,
)
if fig is not None:
fig.tight_layout()
plt.show()
plot_2d_with_bc(custom_domain)
!Warning! Be aware that their is not check or whatsoever to see it the given boundary domain is really the boundary of the given domain.
Square2D domain implements full_bc_domain, which returns a list of SurfacicDomains that represent the all boundary of a square.full_bc_domain.Complement on the mapping¶
Note that the Mapping can take extra arguments:
jacto declare the jacobian of the mapping (if not provided, it is computed using torchtorch.func.jacrev)invto be declared if the mapping is considered invertibleinv_jacfor the jacobian of the inverse (otherwise computed asjac)
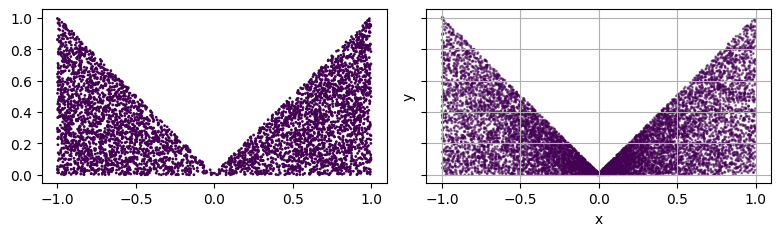
As you should have noticed, once a domain is mapped, the basic sampling will not yield an uniform sampling anymore. To still have it you need to declare an invertible mapping and use sample_post_map.
[14]:
from scimba_torch.integration.monte_carlo import SurfacicSampler, VolumetricSampler
# define a mapping function
def map(x):
"""A simple mapping function."""
return torch.stack([x[:, 0], x[:, 1]*(torch.abs(x[:, 0]) + 1e-2)], dim=1)
# define the inverse mapping function
def inv(y):
"""Inverse of the mapping function."""
return torch.stack([y[:, 0], y[:, 1]/(torch.abs(y[:, 0]) + 1e-2)], dim=1)
# define the mapping
mapping = Mapping(
from_dim=2,
to_dim=2,
map=map,
inv=inv
)
square = simple_square()
# set the mapping for the square domain
square.set_mapping(
map=mapping,
bounds_postmap=[(-1, 1), (0, 1)],
)
# create the sampler
sampler = VolumetricSampler(square)
# sample points
pts = sampler.sample_postmap(5_000)
pts = pts.detach().cpu().numpy()
# plot the points
fig, axs = plt.subplots(1, 2, figsize=(8, 4), sharex=True, sharey=True)
axs[0].scatter(pts[:, 0], pts[:, 1], s=1, c=torch.zeros(pts.shape[0]).numpy())
axs[0].set_aspect('equal')
plot_2d(square, ax=axs[1])
fig.tight_layout()
plt.show()
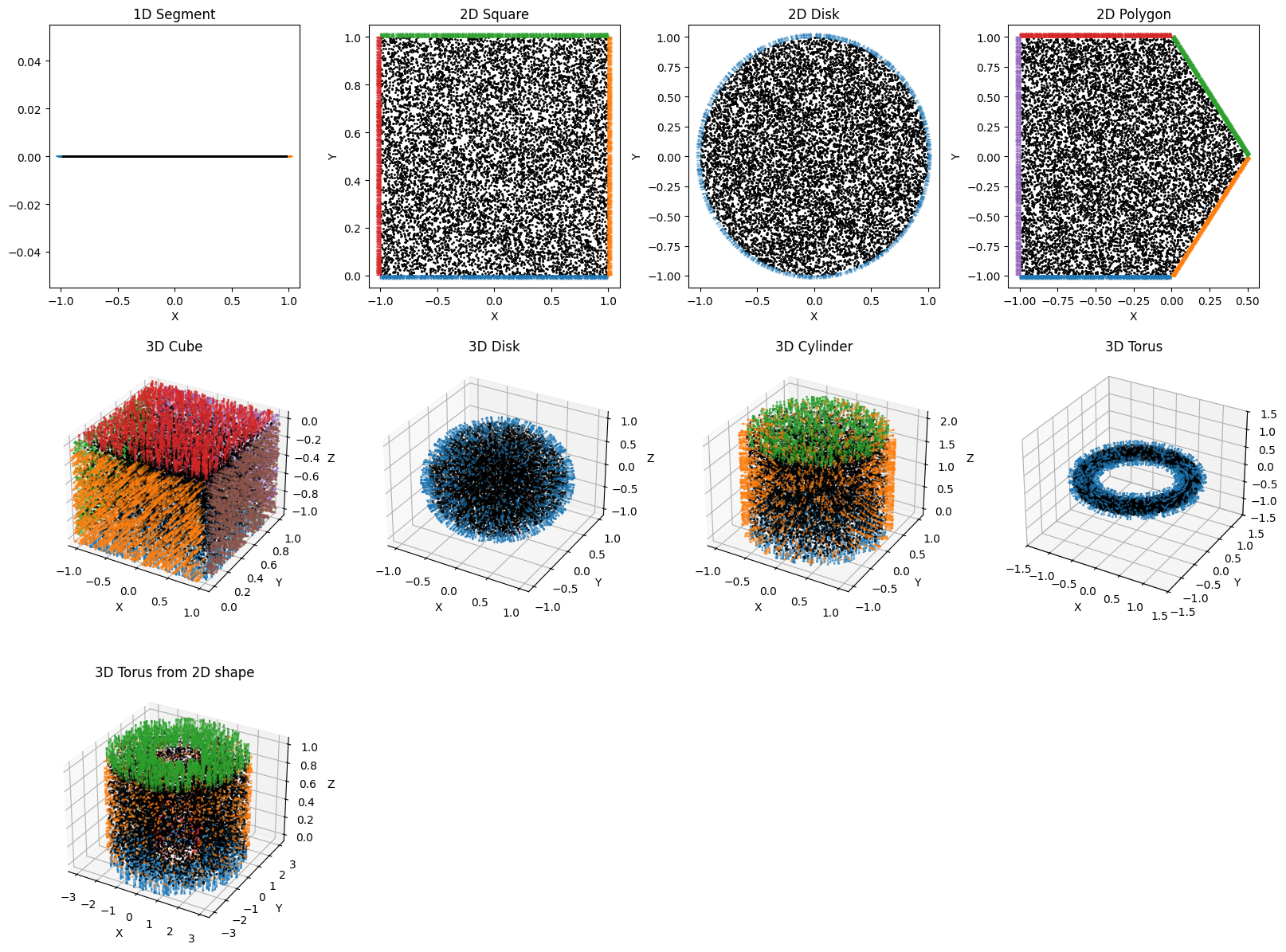
Geometries Showcase¶
[15]:
# 1D domains
segment = Segment1D(
low_high=(-1, 1),
is_main_domain=True
)
# 2D domains
square = Square2D(
bounds=[(-1, 1), (0, 1)],
is_main_domain=True
)
disk = Disk2D(
center=(0, 0),
radius=1,
is_main_domain=True
)
polygon = Polygon2D(
# vertices of the polygon, they should be provided in counter-clockwise order
vertices=[[-1, -1], [0, -1], [.5, 0], [0, 1], [-1, 1]],
is_main_domain=True
)
# 3D domains
cube = Cube3D(
bounds=[(-1, 1), (0, 1), (-1, 0)],
is_main_domain=True
)
disk3d = Disk3D(
center=(0, 0, 0),
radius=1,
is_main_domain=True
)
cylinder = Cylinder3D(
radius=1,
length=2,
is_main_domain=True
)
torus = Torus3D(
center=(0, 0, 0),
radius=1,
tube_radius=0.2,
is_main_domain=True
)
torus_from2d = TorusFrom2DVolume(
base_volume=square,
radius=2,
is_main_domain=True
)
[16]:
# import necessary libraries
from scimba_torch.integration.monte_carlo import VolumetricSampler
# utility function to sample and plot points in a domain
def plot(dict_dom2ax, n=10_000, n_bc=1000):
for domain, ax in dict_dom2ax.items():
# create a sampler for the domain
sampler = VolumetricSampler(domain)
bc_sampler = [SurfacicSampler(bc) for bc in domain.full_bc_domain()]
# sample N points
pts = sampler.sample(n)
bc_pts = [
bc_sampler[i].sample(n_bc, compute_normals=True)
for i in range(len(bc_sampler))
]
# convert to numpy for plotting (& normalize normals)
pts = pts.detach().numpy()
bc_pts = [
(bc, 0.1 * n / torch.linalg.norm(n, axis=-1, keepdims=True))
for bc, n in bc_pts
]
bc_pts = [(bc[0].detach().numpy(), bc[1].detach().numpy()) for bc in bc_pts]
def get_pts(pts):
if domain.dim == 1:
return pts[:, 0], [0] * len(pts)
elif domain.dim == 2:
return pts[:, 0], pts[:, 1]
elif domain.dim == 3:
return pts[:, 0], pts[:, 1], pts[:, 2]
else:
raise ValueError("Domain dimension not supported for plotting.")
ax.scatter(*get_pts(pts), color="k", s=1) # plot the sampled points
for i, (bc, nn) in enumerate(bc_pts):
ax.scatter(
*get_pts(bc), s=1, c=f"C{i}", alpha=0.2
) # plot the boundary points
ax.quiver(
*get_pts(bc), *get_pts(nn), color=f"C{i}", alpha=0.5
) # plot the boundary normals
fig = plt.figure(figsize=(16, 12))
dict_dom2ax = {
segment: fig.add_subplot(3, 4, 1, xlabel="X", title="1D Segment"),
square: fig.add_subplot(3, 4, 2, xlabel="X", ylabel="Y", title="2D Square"),
disk: fig.add_subplot(3, 4, 3, xlabel="X", ylabel="Y", title="2D Disk"),
polygon: fig.add_subplot(3, 4, 4, xlabel="X", ylabel="Y", title="2D Polygon"),
cube: fig.add_subplot(
3, 4, 5, projection="3d", xlabel="X", ylabel="Y", zlabel="Z", title="3D Cube"
),
disk3d: fig.add_subplot(
3, 4, 6, projection="3d", xlabel="X", ylabel="Y", zlabel="Z", title="3D Disk"
),
cylinder: fig.add_subplot(
3,
4,
7,
projection="3d",
xlabel="X",
ylabel="Y",
zlabel="Z",
title="3D Cylinder",
),
torus: fig.add_subplot(
3,
4,
8,
projection="3d",
xlabel="X",
ylabel="Y",
zlabel="Z",
xlim=(-1.5, 1.5),
ylim=(-1.5, 1.5),
zlim=(-1.5, 1.5),
title="3D Torus",
),
torus_from2d: fig.add_subplot(
3,
4,
9,
projection="3d",
xlabel="X",
ylabel="Y",
zlabel="Z",
title="3D Torus from 2D shape",
),
}
# plot the domains
plot(dict_dom2ax)
fig.tight_layout()
plt.show()
[ ]: