Scimba basics II: Natural gradient preconditioners¶
In the previous tutorials, we used a PINN fit through a natural gradient descent algorithm to the solution of a 2D laplacian problem.
Scimba implements two natural gradient preconditioners for gradient descent algorithms: Energy Natural Gradient (ENG) and Anagram.
Both can be used for PINNs and the task of projecting a function onto an approximation space.
This tutorial demonstrates further the use of ENG and Anagram preconditioners in Scimba for PINNs on the 2D laplacian example, and for projectors. A following tutorial explains how to model a pde so that it can be used with a natural gradient preconditioned PINN.
Let us first define the problem we consider, and solve it with a not preconditioned PINN for the reference.
Approximation with a not preconditioned PINN¶
[1]:
import torch
from scimba_torch.approximation_space.nn_space import NNxSpace
from scimba_torch.domain.meshless_domain.domain_2d import Square2D
from scimba_torch.integration.monte_carlo import DomainSampler, TensorizedSampler
from scimba_torch.integration.monte_carlo_parameters import UniformParametricSampler
from scimba_torch.neural_nets.coordinates_based_nets.mlp import GenericMLP
from scimba_torch.physical_models.elliptic_pde.laplacians import (
Laplacian2DDirichletStrongForm,
)
from scimba_torch.utils.scimba_tensors import LabelTensor
torch.manual_seed(0)
domain_x = Square2D([(0.0, 1), (0.0, 1)], is_main_domain=True)
domain_mu = [(0.5, 1.0)]
sampler = TensorizedSampler(
[DomainSampler(domain_x), UniformParametricSampler(domain_mu)]
)
def f_rhs(xs: LabelTensor, ms: LabelTensor) -> torch.Tensor:
x, y = xs.get_components()
mu = ms.get_components()
pi = torch.pi
return 2 * (2.0 * mu * pi) ** 2 * torch.sin(2.0 * pi * x) * torch.sin(2.0 * pi * y)
def f_bc(xs: LabelTensor, ms: LabelTensor) -> torch.Tensor:
x, _ = xs.get_components()
return x * 0.0
space = NNxSpace(
1, # the output dimension
1, # the parameter's space dimension
GenericMLP, # the type of neural network
domain_x, # the geometric domain
sampler, # the sampler
layer_sizes=[16, 16], # the size of intermediate layers
)
pde = Laplacian2DDirichletStrongForm(space, f=f_rhs, g=f_bc)
We define a PinnsElliptic object to approximate the solution of pde.
Notice the use of keyword argument bc_weight to scale the loss induced by the boundary condition relatively to the loss of the residual inside the domain.
[2]:
from scimba_torch.numerical_solvers.elliptic_pde.pinns import PinnsElliptic
pinn = PinnsElliptic(pde, # the pde to be solved
bc_type="weak", # use explicitly the boundary conditions
bc_weight=50.) # the weight of the loss of the boundary condition
The next step is to solve the PINN, that is to find the parameters of the approximation space (here a multi-layer perceptron) to minimize the residual inside the domain and the boundary condition, here Dirichlet.
At each step, this requires to sample the interior and the boundaries of the considered domain \(\Omega\). Below, we time 1000 steps of the Adam optimizer.
[3]:
import timeit
start = timeit.default_timer()
pinn.solve(epochs=1000, #number of steps of optimization
n_collocation=1000, #number of sample points inside the domain
n_bc_collocation=1000, #number of sample points on the boundary
verbose=False)
end = timeit.default_timer()
print("time in approximation with a PINN: ", end - start)
print("best loss: ", pinn.best_loss)
Training: 100%|||||||||||||||| 1000/1000[00:04<00:00] , loss: 5.6e+02 -> 1.8e+01
time in approximation with a PINN: 5.348011291993316
best loss: 18.456674964641266
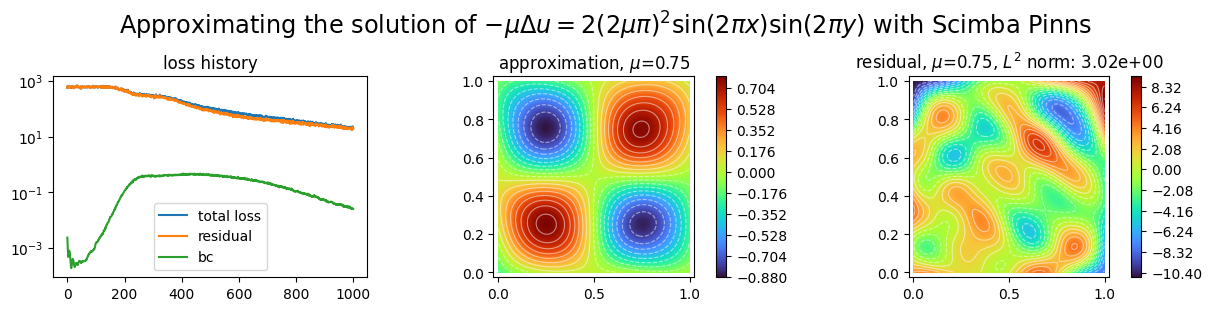
Let us plot the obtained approximation, the residual inside the domain and the history of the loss variation during the optimization.
[4]:
import matplotlib.pyplot as plt
from scimba_torch.plots.plots_nd import plot_abstract_approx_spaces
plot_abstract_approx_spaces(
pinn.space, # the approximation space
domain_x, # the spatial domain
domain_mu, # the parameter's domain
loss=pinn.losses, # optional plot of the loss: the losses
residual=pinn.pde, # optional plot of the residual of the pde
draw_contours=True, # plotting isolevel lines
n_drawn_contours=20, # number of isolevel lines,
title=(
"Approximating the solution of "
r"$- \mu\Delta u = 2(2\mu\pi)^2 \sin(2\pi x) \sin(2\pi y)$"
" with Scimba Pinns"
),
)
plt.show()
Approximation with preconditioned PINNs¶
Scimba implements two natural gradient descent algorithms for PINNs: Energy Natural Gradient and Anagram.
For linear PDEs (as is the laplacian equation under consideration), both approaches rely on pseudo inverting the so called energy Gram matrix, however the numerical methods used to do so differ.
Energy Natural Gradient preconditioning¶
[5]:
ENGspace = NNxSpace(
1, # the output dimension
1, # the parameter's space dimension
GenericMLP, # the type of neural network
domain_x, # the geometric domain
sampler, # the sampler
layer_sizes=[16, 16], # the size of intermediate layers
)
ENGpde = Laplacian2DDirichletStrongForm(ENGspace, f=f_rhs, g=f_bc)
PINNs with ENG descent algorithm are NaturalGradientPinnsElliptic objects.
Notice the use of the matrix_regularization argument when instantiating the pinn. It is set to \(10^{-6}\) by default. In your examples, play around with this parameter as preconditioning with ENG is sensitive to this parameter.
[6]:
from scimba_torch.numerical_solvers.elliptic_pde.pinns import (
NaturalGradientPinnsElliptic,
)
ENGpinn = NaturalGradientPinnsElliptic(ENGpde,
bc_type="weak",
bc_weight=50.,
matrix_regularization=1e-6)
Use SGD algorithm together with a line search for adaptive choice of learning rate.
[7]:
start = timeit.default_timer()
ENGpinn.solve(epochs=200,
n_collocation=1000,
n_bc_collocation=1000,
verbose=False)
end = timeit.default_timer()
print("time in approximation with ENG preconditioned PINN: ", end - start)
print("best loss: ", ENGpinn.best_loss)
Training: 100%|||||||||||||||||| 200/200[00:04<00:00] , loss: 6.0e+02 -> 1.0e-04
time in approximation with ENG preconditioned PINN: 4.557641125051305
best loss: 9.994664816575404e-05
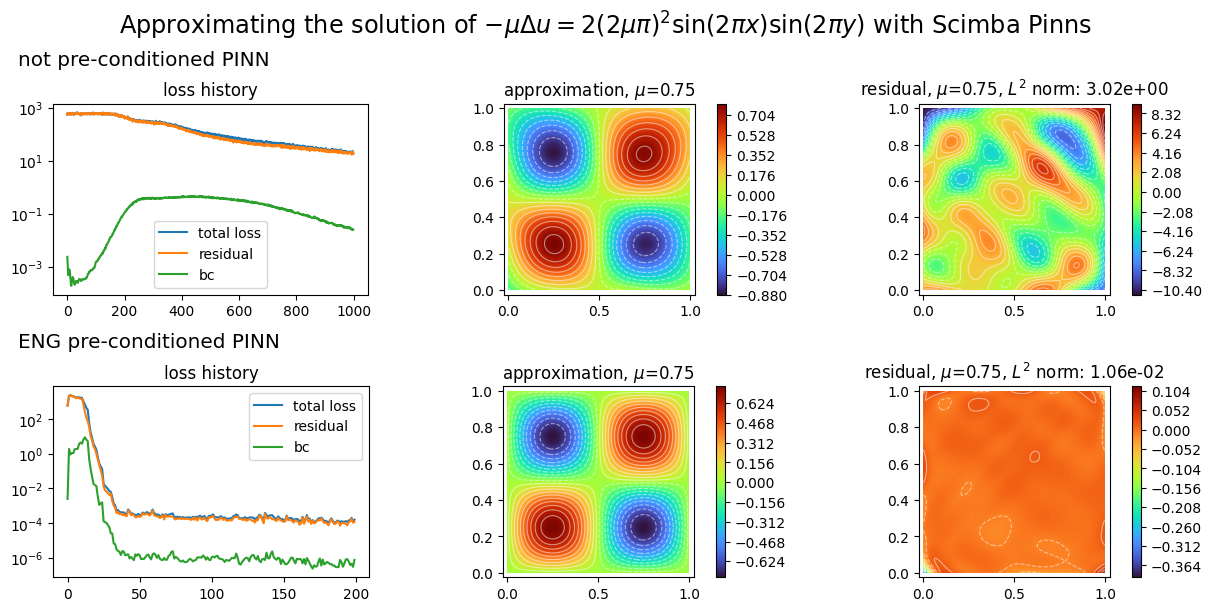
We plot the two PINNs with:
[8]:
plot_abstract_approx_spaces(
(pinn.space, ENGpinn.space), # the approximation spaces
domain_x, # the spatial domain
domain_mu, # the parameter's domain
loss=(pinn.losses, ENGpinn.losses), # optional plot of the loss: the losses
residual=(pinn.pde, ENGpinn.pde), # optional plot of the residual of the pde
draw_contours=True, # plotting isolevel lines
n_drawn_contours=20, # number of isolevel lines,
title=(
"Approximating the solution of "
r"$- \mu\Delta u = 2(2\mu\pi)^2 \sin(2\pi x) \sin(2\pi y)$"
" with Scimba Pinns"
),
titles=("not preconditioned PINN", "ENG preconditioned PINN"),
)
plt.show()
Anagram preconditioning¶
In order to demonstrate Anagram use, we define new approximation space and PDE:
[9]:
ANAspace = NNxSpace(
1, # the output dimension
1, # the parameter's space dimension
GenericMLP, # the type of neural network
domain_x, # the geometric domain
sampler, # the sampler
layer_sizes=[16, 16], # the size of intermediate layers
)
ANApde = Laplacian2DDirichletStrongForm(ANAspace, f=f_rhs, g=f_bc)
PINNs with Anagram gradient descent algorithm are AnagramPinnsElliptic objects.
Notice the use below of the svd_threshold argument when creating the AnagramPinnsElliptic object. The success of the optimization is very sensitive to this parameter; its default value of \(10^{-6}\) might be a very bad choice for some examples.
[10]:
from scimba_torch.numerical_solvers.elliptic_pde.pinns import AnagramPinnsElliptic
ANApinn = AnagramPinnsElliptic(
ANApde, bc_type="weak", bc_weight=50.0, svd_threshold=0.1
)
start = timeit.default_timer()
ANApinn.solve(epochs=200, n_collocation=1000, n_bc_collocation=1000, verbose=False)
end = timeit.default_timer()
print("time in approximation with Anagram preconditioned PINN: ", end - start)
print("best loss: ", ANApinn.best_loss)
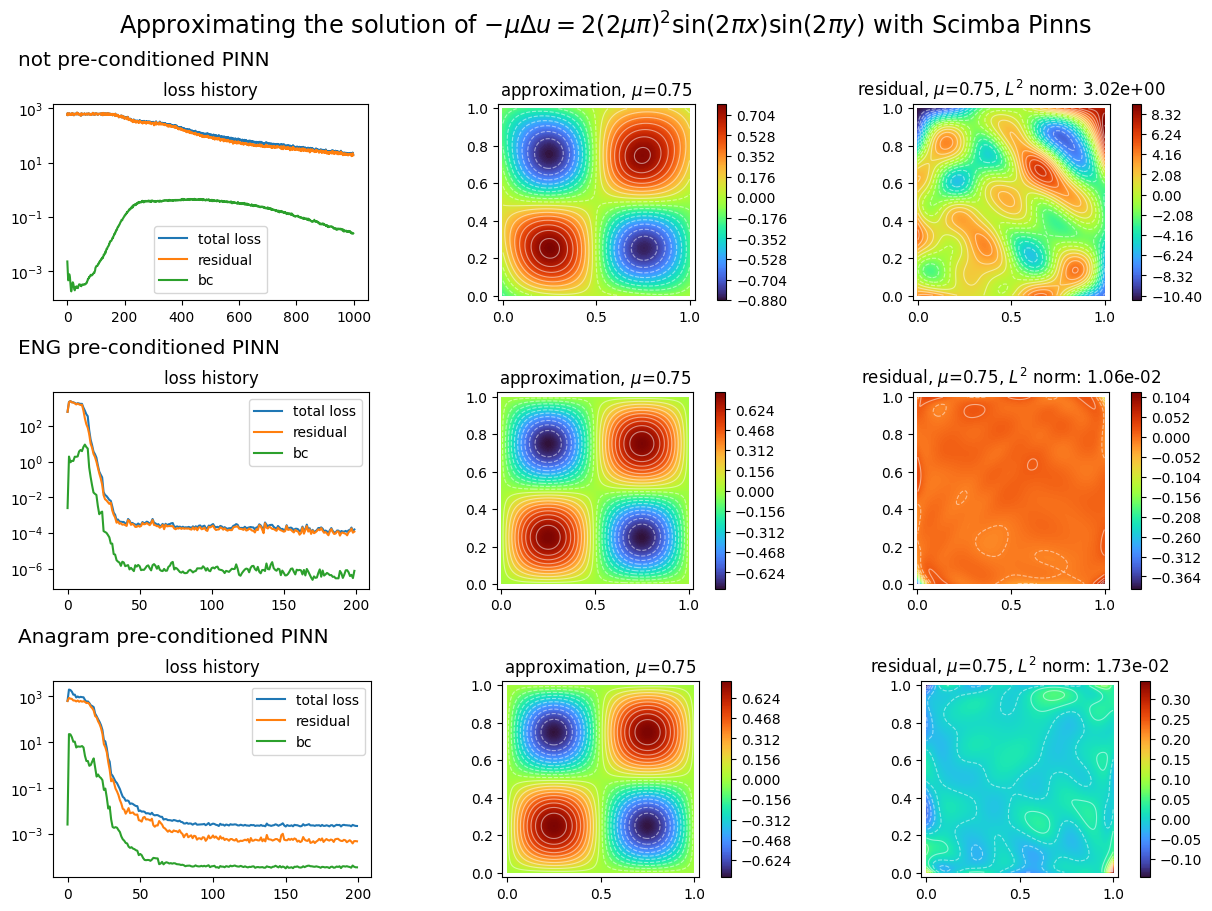
plot_abstract_approx_spaces(
(pinn.space, ENGpinn.space, ANApinn.space), # the approximation space
domain_x, # the spatial domain
domain_mu, # the parameter's domain
loss=(
pinn.losses,
ENGpinn.losses,
ANApinn.losses,
), # optional plot of the loss: the losses
residual=(
pinn.pde,
ENGpinn.pde,
ANApinn.pde,
), # optional plot of the residual of the pde
draw_contours=True, # plotting isolevel lines
n_drawn_contours=20, # number of isolevel lines,
title=(
"Approximating the solution of "
r"$- \mu\Delta u = 2(2\mu\pi)^2 \sin(2\pi x) \sin(2\pi y)$"
" with Scimba Pinns"
),
titles=(
"not preconditioned PINN",
"ENG preconditioned PINN",
"Anagram preconditioned PINN",
),
)
plt.show()
Training: 100%|||||||||||||||||| 200/200[00:10<00:00] , loss: 6.3e+02 -> 2.1e-03
time in approximation with Anagram preconditioned PINN: 10.534930417023133
best loss: 0.002102398089162228
We explain in this tutorial how to write a model for which natural gradient preconditioners can be used.
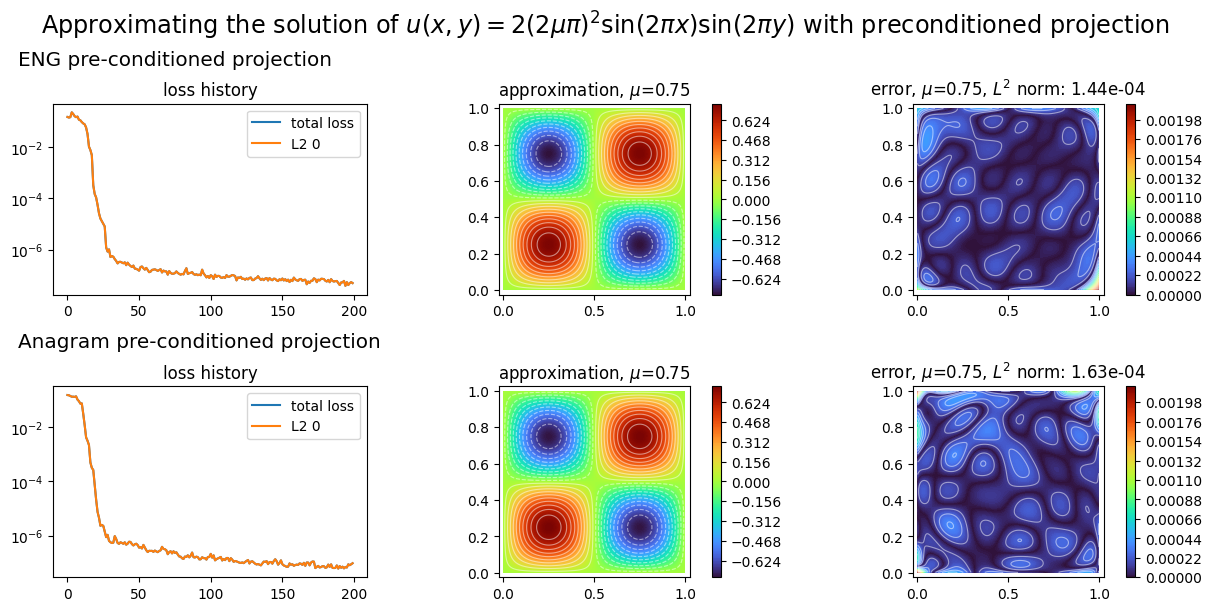
preconditioned projectors¶
To finish with, we show how to project a known function on an approximation space with preconditioned gradient descent optimization. We project the exact solution the 2D laplacian problem considered so far.
[11]:
from scimba_torch.numerical_solvers.collocation_projector import (
NaturalGradientProjector,
)
def exact_sol(xs: LabelTensor, ms: LabelTensor) -> torch.Tensor:
x, y = xs.get_components()
mu = ms.get_components()
pi = torch.pi
return mu * torch.sin(2.0 * pi * x) * torch.sin(2.0 * pi * y)
ENGspace = NNxSpace(1, 1, GenericMLP, domain_x, sampler, layer_sizes=[16, 16])
ENGprojector = NaturalGradientProjector(ENGspace, exact_sol)
start = timeit.default_timer()
ENGprojector.solve(epochs=200, n_collocation=2000, verbose=False)
end = timeit.default_timer()
print("time in projection with ENG preconditioning: ", end - start)
print("best L2 error: ", ENGprojector.best_loss)
ANAspace = NNxSpace(1, 1, GenericMLP, domain_x, sampler, layer_sizes=[16, 16])
ANAprojector = NaturalGradientProjector(ANAspace, exact_sol)
start = timeit.default_timer()
ANAprojector.solve(epochs=200, n_collocation=2000, verbose=False)
end = timeit.default_timer()
print("time in projection with Anagram preconditioning: ", end - start)
print("best L2 error: ", ANAprojector.best_loss)
Training: 100%|||||||||||||||||| 200/200[00:01<00:00] , loss: 1.5e-01 -> 3.8e-08
time in projection with ENG preconditioning: 1.680992542009335
best L2 error: 3.799798007426025e-08
Training: 100%|||||||||||||||||| 200/200[00:01<00:00] , loss: 1.6e-01 -> 6.0e-08
time in projection with Anagram preconditioning: 1.6535701660322957
best L2 error: 6.021761856087472e-08
[12]:
plot_abstract_approx_spaces(
(ENGprojector.space, ANAprojector.space), # the approximation space
domain_x, # the spatial domain
domain_mu, # the parameter's domain
# optional plot of the loss: the losses
loss=(ENGprojector.losses, ANAprojector.losses),
error=exact_sol, # plot the error w.r.t. the exact solution
draw_contours=True, # plotting isolevel lines
n_drawn_contours=20, # number of isolevel lines,
title=(
"Approximating the solution of "
r"$u(x,y) = 2(2\mu\pi)^2 \sin(2\pi x) \sin(2\pi y)$"
" with preconditioned projection"
),
titles=("ENG preconditioned projection", "Anagram preconditioned projection"),
)
plt.show()
[ ]: