Projection of a function on an approximation space.¶
We demonstrate here how to approximate a known function with a neural network in Scimba on 2 examples. The function to be approximated has to be known as a procedure for its evaluation.
First example: highly curved function¶
Let
where:
Let us import the required libraries and modules then implement the function to approximate:
[1]:
import matplotlib.pyplot as plt
import torch
from scimba_torch.utils.scimba_tensors import LabelTensor
def func(x: LabelTensor, mu: LabelTensor):
x1, x2 = x.get_components()
r = torch.sqrt(x1**2 + x2**2)
theta = torch.atan(x2 / x1)
r = r + 0.05 * torch.cos(4 * torch.pi * theta)
sigma_theta = 0.53
sigma_r = 0.008
return (
torch.sin(theta)
* torch.cos(theta)
* torch.exp(-((theta - torch.pi / 4) ** 2) / (2 * sigma_theta**2.0))
* torch.exp(-((r - 0.5) ** 2) / (2 * sigma_r**2.0))
) + 1.1 * (
torch.sin(1.5 * theta)
* torch.cos(0.5 * theta)
* torch.exp(-0.5 * ((theta - torch.pi / 4) ** 2) / (2 * sigma_theta**2.0))
* torch.exp(-1.25 * ((r - 0.5) ** 2) / (2 * sigma_r**2.0))
)
We will approximate this function on the domain \(\Omega = [0, 1] \times [0,1]\):
[2]:
from scimba_torch.domain.meshless_domain.domain_2d import Square2D
from scimba_torch.integration.monte_carlo import DomainSampler, TensorizedSampler
from scimba_torch.integration.monte_carlo_parameters import UniformParametricSampler
omega = Square2D([(0, 1), (0, 1)], is_main_domain=True)
sampler = TensorizedSampler([DomainSampler(omega), UniformParametricSampler([])])
The base method: projection on a generic MLP based approximation space¶
We first do as if we had no knowledge of the function to approximate and search an approximation on a NN with a generic MLP architecture:
[3]:
from scimba_torch.approximation_space.nn_space import NNxSpace
from scimba_torch.neural_nets.coordinates_based_nets.mlp import GenericMLP
space1 = NNxSpace(1, 0, GenericMLP, omega, sampler, layer_sizes=[18, 18])
The parameters of the NN have to be tuned to get an approximation of the desired function; this is what we call projection on the approximation space. To this end, we define a so-called projector:
[4]:
from scimba_torch.numerical_solvers.collocation_projector import NaturalGradientProjector
p1 = NaturalGradientProjector(space1, func)
Here we use a projector with natural gradient preconditioning, i.e. with natural gradient preconditioned gradient descent. The projection is computed by calling the solve method of the projector:
[5]:
p1.solve(epochs=400, n_collocation=10000)
Training: 100%|||||||||||||||||| 400/400[00:12<00:00] , loss: 1.1e-02 -> 2.5e-06
Plot the approximation with:
[6]:
import matplotlib.pyplot as plt
from scimba_torch.plots.plots_nd import plot_abstract_approx_spaces
plot_abstract_approx_spaces(
p1.space,
omega,
loss=p1.losses,
solution=func,
error=func,
draw_contours=True,
n_drawn_contours=20,
title="Projection with Energy Natural Gradient",
)
plt.show()
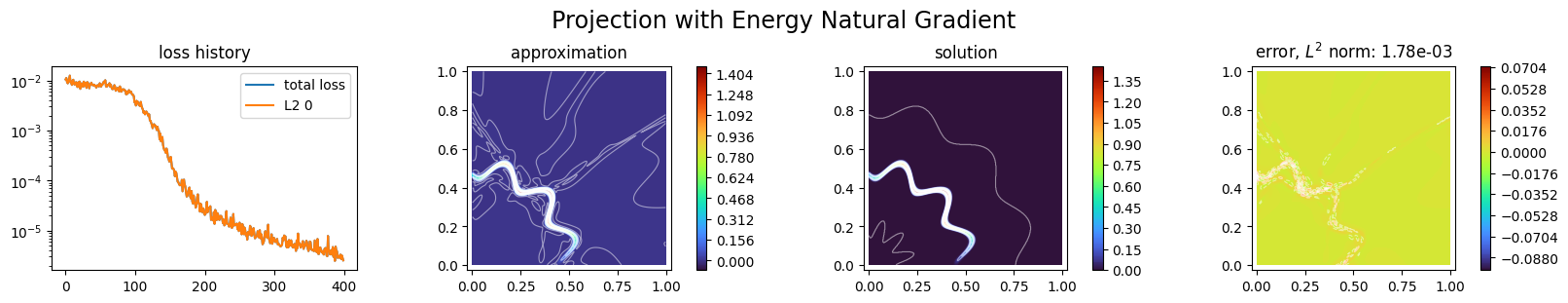
This approach suits well to the case where the function to be approximated is given as a black-box function.
Adapt the network architecture to the function to approximate¶
In the present case where we know a formula for the function to be approximated, we can take advantage of it.
Remarking that the function we want to approximate is essentially a function of \(\theta\) and \(r\) where:
we propose to apply a change of variable to feed the network with \(\theta\) and \(r\). To this end, we apply a pre-processing to the input of the approximation space:
[7]:
def variable_change(x, mu):
x1, x2 = x.get_components()
r = torch.sqrt(x1**2 + x2**2)
theta = torch.atan2(x2, x1)
r = r + 0.05 * torch.cos(4 * torch.pi * theta)
return torch.cat([r, theta], dim=1)
space2 = NNxSpace(1, 0, GenericMLP, omega, sampler, layer_sizes=[18, 18], pre_processing=variable_change)
p2 = NaturalGradientProjector(space2, func)
p2.solve(epochs=200, n_collocation=10000)
Training: 100%|||||||||||||||||| 200/200[00:06<00:00] , loss: 2.8e-02 -> 4.1e-08
[8]:
plot_abstract_approx_spaces(
p2.space,
omega,
loss=p2.losses,
solution=func,
error=func,
draw_contours=True,
n_drawn_contours=20,
title="Projection with Energy Natural Gradient and change of variable",
)
plt.show()
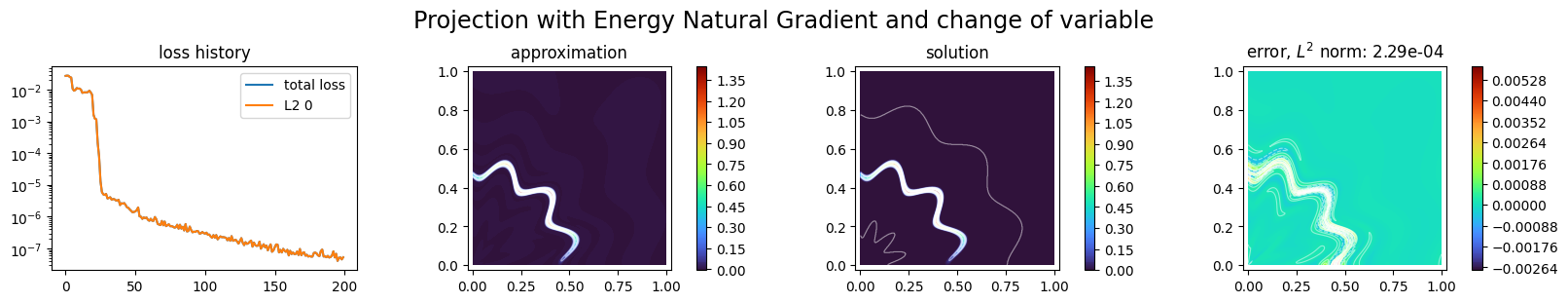
Finally, since the function to approximate is the sum of two quantities, we try to compute its approximation with to MLP-like NNs which outputs are gathered; this can be done with a SeparatedNNxSpace approximation space:
[9]:
from scimba_torch.approximation_space.nn_space import SeparatedNNxSpace
space3 = SeparatedNNxSpace(
1,
0,
2, # the number of NNs
GenericMLP,
omega,
sampler,
layer_sizes=[12, 12],
pre_processing=variable_change,
)
p3 = NaturalGradientProjector(space3, func)
p3.solve(epochs=200, n_collocation=10000)
Training: 100%|||||||||||||||||| 200/200[00:16<00:00] , loss: 1.4e-02 -> 4.3e-07
[10]:
plot_abstract_approx_spaces(
p3.space,
omega,
loss=p3.losses,
solution=func,
error=func,
draw_contours=True,
n_drawn_contours=20,
title="Projection with Energy Natural Gradient and change of variable on a separable space",
)
plt.show()
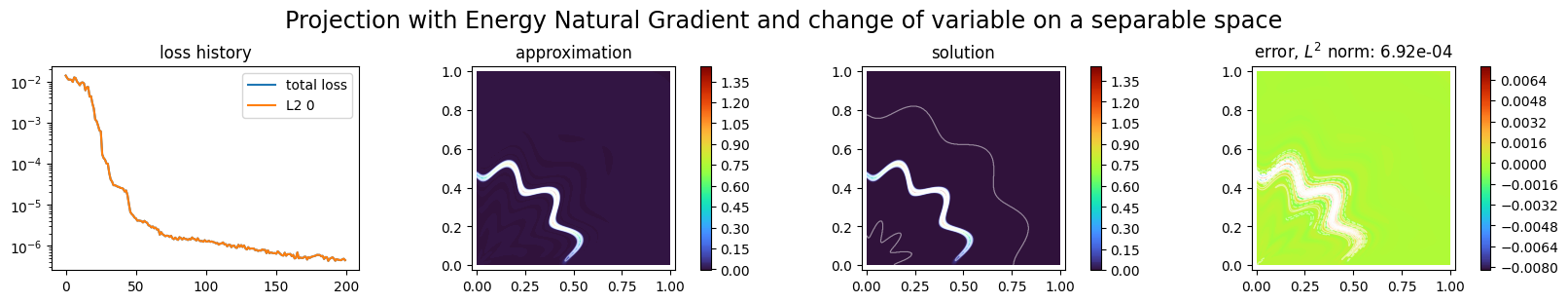
Second example: learning an oscillatory function¶
We now consider the function:
where \({\bf x}=(x, y)\) and \(\sigma = 0.4\).
We want to learn \(f\) on \(\Omega = [-1, 1]\times [-1, 1]\).
[11]:
def func(xy: LabelTensor, mu: LabelTensor):
x, y = xy.get_components()
sigma = 0.4
return (
torch.sin(2.5 * torch.pi * x)
* torch.sin(4.5 * torch.pi * y)
* torch.exp(-((x) ** 2 + (y) ** 2) / (2 * sigma**2))
)
omega = Square2D([(-1.0, 1.0), (-1.0, 1.0)], is_main_domain=True)
sampler = TensorizedSampler([DomainSampler(omega), UniformParametricSampler([])])
The base method: projection on a generic MLP based approximation space¶
As previously, we first do as if we had no knowledge on the function to approximate, and project it on a MLP like network:
[12]:
torch.manual_seed(0)
space1 = NNxSpace(1, 0, GenericMLP, omega, sampler, layer_sizes=[18, 18])
p1 = NaturalGradientProjector(space1, func)
p1.solve(epochs=400, n_collocation=10000)
Training: 100%|||||||||||||||||| 400/400[00:11<00:00] , loss: 4.0e-02 -> 4.2e-07
Remark: the training is a stochastic process; try to change the seed if it does not improve the loss.
[13]:
plot_abstract_approx_spaces(
p1.space,
omega,
loss=p1.losses,
solution=func,
error=func,
draw_contours=True,
n_drawn_contours=20,
title="Projection with Energy Natural Gradient",
)
plt.show()
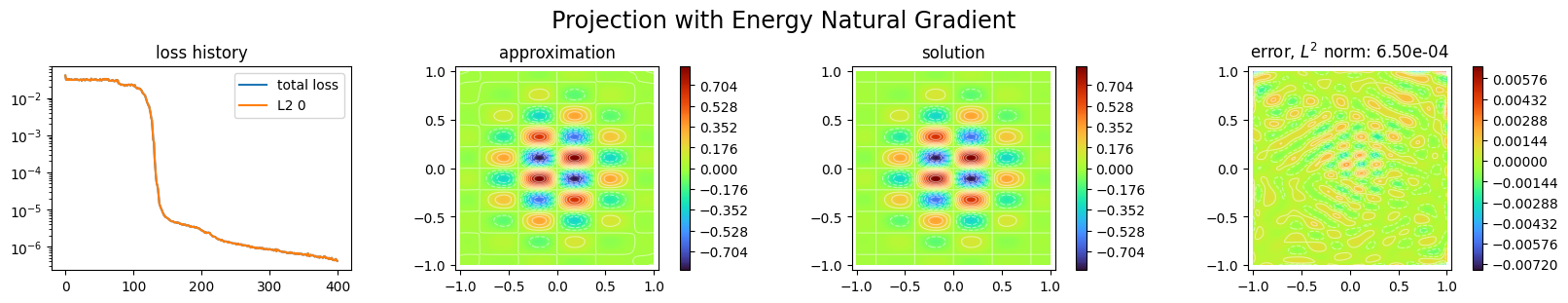
Adapt the network architecture to the function to approximate¶
Let us now project the function on a space with Fourier modes. Such an approximation space can be instantiated with:
[14]:
from scimba_torch.approximation_space.spectral_space import SpectralxSpace
space2 = SpectralxSpace(
1, # nb of unknowns
"sine", # basis functions
6, # nb of modes per direction
omega.bounds, # domain bounds
sampler # sampler
)
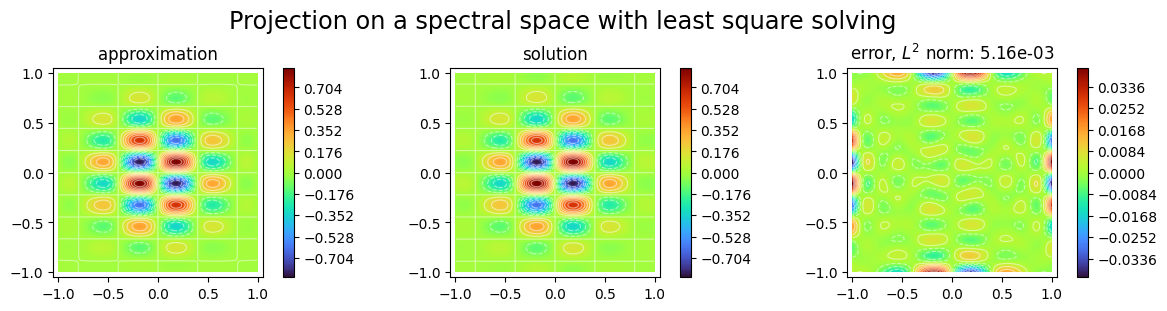
We first propose to adjust the parameters with a least squares solver:
[15]:
from scimba_torch.numerical_solvers.collocation_projector import LinearProjector
p2 = LinearProjector(space2, func)
p2.solve(n_collocation=10000, verbose=True)
Training done!
Final loss value: 2.608e-05
Best loss value: 2.850e-05
[16]:
plot_abstract_approx_spaces(
p2.space,
omega,
solution=func,
error=func,
draw_contours=True,
n_drawn_contours=20,
title="Projection on a spectral space with least square solving",
)
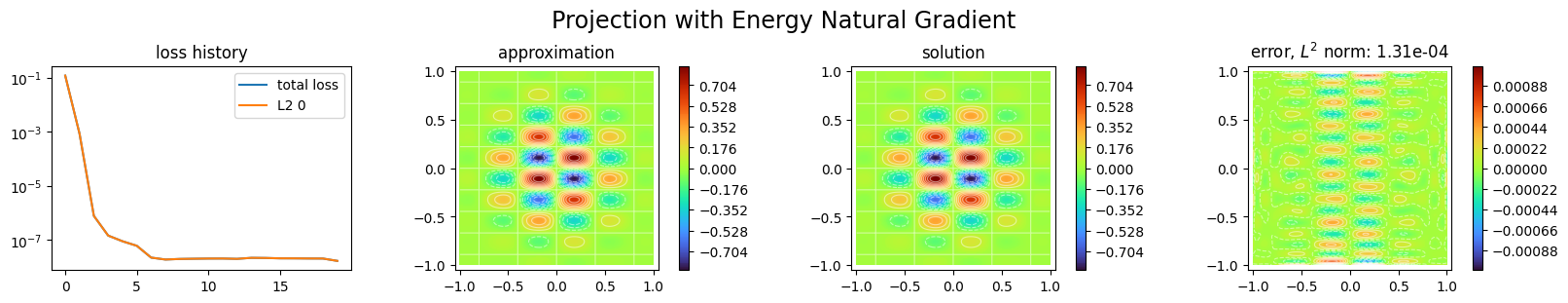
We finally project the function with Natural Gradient preconditioning:
[17]:
space3 = SpectralxSpace(
1, # nb of unknowns
"sine", # basis functions
6, # nb of modes per direction
omega.bounds, # domain bounds
sampler # sampler
)
p3 = NaturalGradientProjector(space3, func)
p3.solve(epochs=20, n_collocation=10000)
Training: 100%|||||||||||||||||||| 20/20[00:00<00:00] , loss: 1.2e-01 -> 1.7e-08
[18]:
plot_abstract_approx_spaces(
p3.space,
omega,
loss=p3.losses,
solution=func,
error=func,
draw_contours=True,
n_drawn_contours=20,
title="Projection with Energy Natural Gradient",
)
plt.show()
[ ]: